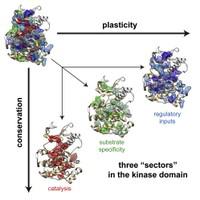
 Proteins can fold
spontaneously into well-defined three-dimensional structures and can carry out
complex biochemical reactions such as binding, catalysis, and long-range information
transfer. The precision required for these properties is achieved while also
preserving evolvability – the capacity to adapt in response to fluctuating
selection pressures in the environment. What is the basic design of proteins
that supports all of these properties? Recent work suggests that rather than
direct physical analysis, statistical analysis of genome sequences provides a
powerful and general approach to this problem. Using different methodologies,
this approach has revealed both direct structural contacts as well as collective
functional modes within protein structures. In this talk, I will present the
current state of these approaches and the possibility of unifying them into a
single theoretical framework for representing the evolutionary design of
proteins.
Proteins can fold
spontaneously into well-defined three-dimensional structures and can carry out
complex biochemical reactions such as binding, catalysis, and long-range information
transfer. The precision required for these properties is achieved while also
preserving evolvability – the capacity to adapt in response to fluctuating
selection pressures in the environment. What is the basic design of proteins
that supports all of these properties? Recent work suggests that rather than
direct physical analysis, statistical analysis of genome sequences provides a
powerful and general approach to this problem. Using different methodologies,
this approach has revealed both direct structural contacts as well as collective
functional modes within protein structures. In this talk, I will present the
current state of these approaches and the possibility of unifying them into a
single theoretical framework for representing the evolutionary design of
proteins.

