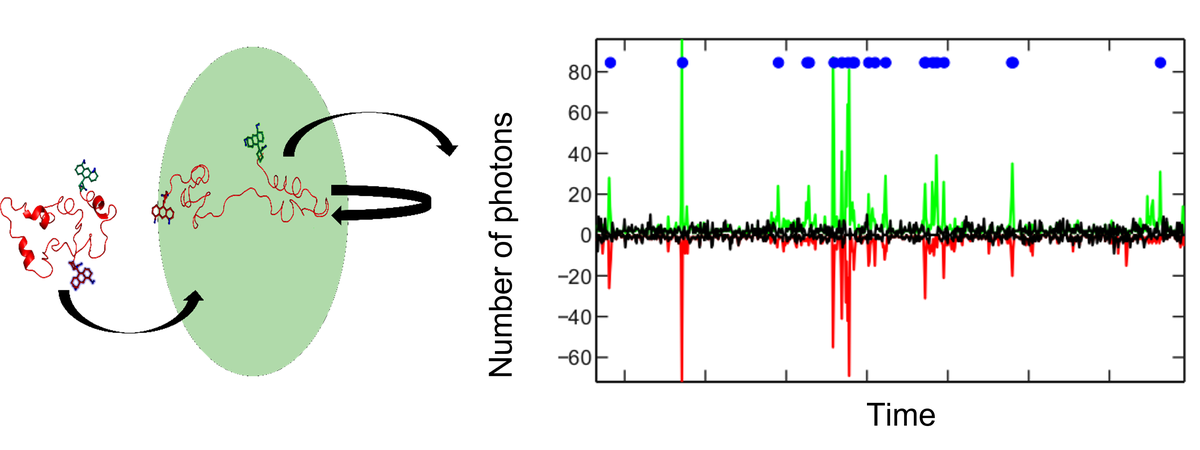
Biological processes are intrinsically dynamic, which dictates many key aspects of protein function. Intrinsically disordered proteins (IDPs) are an extreme case of fluctuating 3D folds (conformations), which makes their characterization challenging. My lab specializes in multidimensional single-molecule fluorescence spectroscopy, a uniquely suited technique to resolve microscopic distributions of states and dynamics. In this talk, I will share some of our recent single-molecule data of two prototypical IDPs, Sic1 and 4E-BP2, which form “fuzzy” complexes with their partners. Conformational ensembles of these proteins were estimated from a variety of experimental restraints, such as nuclear magnetic resonance, small-angle X-ray scattering and single-molecule fluorescence. Data mining of these ensembles reveals features that deviate from polymer physics, which may result from evolutionary pressure for specific biological functions. An integrative use of multiple biophysical experiments probing disparate scales, computational modelling and polymer physics provides valuable insights into IDPs and their diverse biological functions.
Function without Structure: Single-molecule Experiments and Modelling of Disordered Proteins
Host: Josh Milstein